YADA 3.0 Tutorial
 YADA 3.0 is the newest version of our Yet Another Deconvolution Algorithm
(Y.A.D.A) tool. It can perform lighting fast deconvolution, with accurate
results.
YADA 3.0 is the newest version of our Yet Another Deconvolution Algorithm
(Y.A.D.A) tool. It can perform lighting fast deconvolution, with accurate
results.
Please cite:
XXXXX
Hyperlinks
How to Use (Command Line Version)
YADA 3.0 Algorithm
The algorithm begins by locating all
sets of peaks that are within an acceptable m/z range satisfying an
envelope of a given charge state; so, for example, a 2+ envelope would be
assumed if consecutive peaks are ~0.5 m/z apart. The minimum number of consecutive peaks and minimum
peak intensity are user defined variables.
In what follows, for each envelope candidate, a normalized dot product
is performed between the experimental and a theoretical profile of intensities.
YADA generates the theoretical profile by resorting to an array of spline
regressors that have been trained with the averagine
model (https://doi.org/10.1016/1044-0305(95)00017-8); this array can model up
to 25 envelope peaks. Subsequently, YADA applies a maximum parsimony filter
that will guarantee that each mass spectral peak belongs to only one isotopic
envelope candidate; envelopes are chosen according to their highest scoring dot
product. Finally, only envelopes with
dot product above a given threshold are reported to the user.
How to Use (GUI Version)
1. Installation
The latest
version of YADA 3.0 can be found here. It requires
.NET 6.0 or higher to run, which can be downloaded here.
An example
raw file can be found here.
2. Deconvolution Parameters
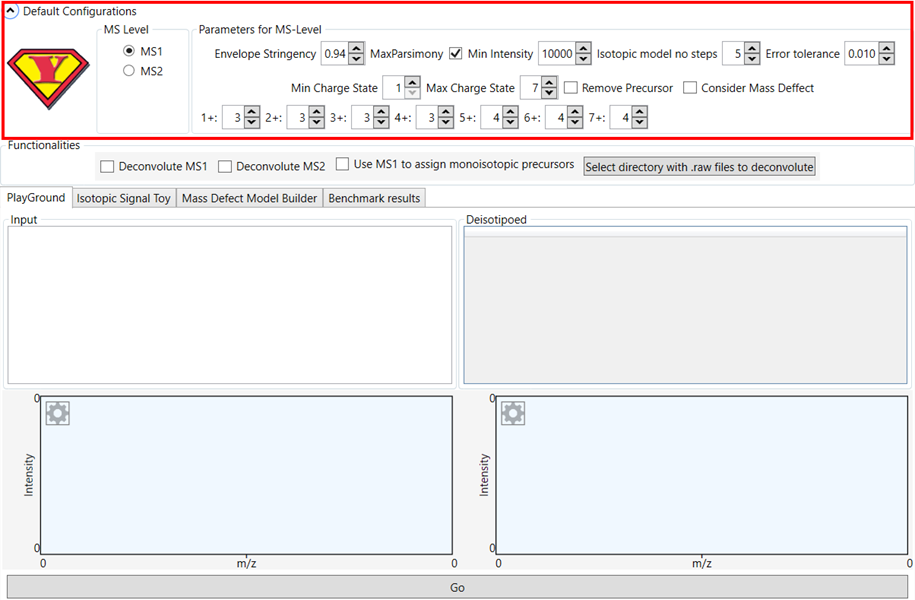
Figure 1 - YADA 3.0 start screen.
YADA 3.0 provides
several configurable deconvolution parameters, which can be changed in the
graphical user interface (GUI). The MS Level box allows configuring the
parameters for MS1 and MS2 spectra deconvolution separately.
Default
parameters area already configured for optimal performance in most common
proteomics experiments in high-resolution data.
A brief
description of each parameter:
· Envelope Stringency: Minimum isotopic
envelope similarity score required to accept an envelope.
· MaxParsimony: Whether to allow an ion to belong
to two different isotopic envelope. If checked, the
lower scoring envelope for each ion is ignored.
· Min Intensity: Minimum required ion
intensity required to consider an ion.
· Isotopic Model No Steps: Number of
isotopes to be modelled for each ion.
· Error Tolerance: Mass error allowed.
Measured in Daltons.
· Min Charge State: Minimum charge
state to consider in the spectrum.
· Max Charge State: Maximum charge
state to consider in the spectrum.
· Remove Precursor: If selected,
envelopes matching the precursor mass will be ignored.
· Consider Mass Defect: If selected,
isotopic envelopes that do not fit YADA’s mass defect model will be ignored.
· +1, +2, +3… :
Minimum number of ions required to accept an isotopic envelope.
3. Deconvoluting Thermo
.raw / .ms1 and .ms2 files
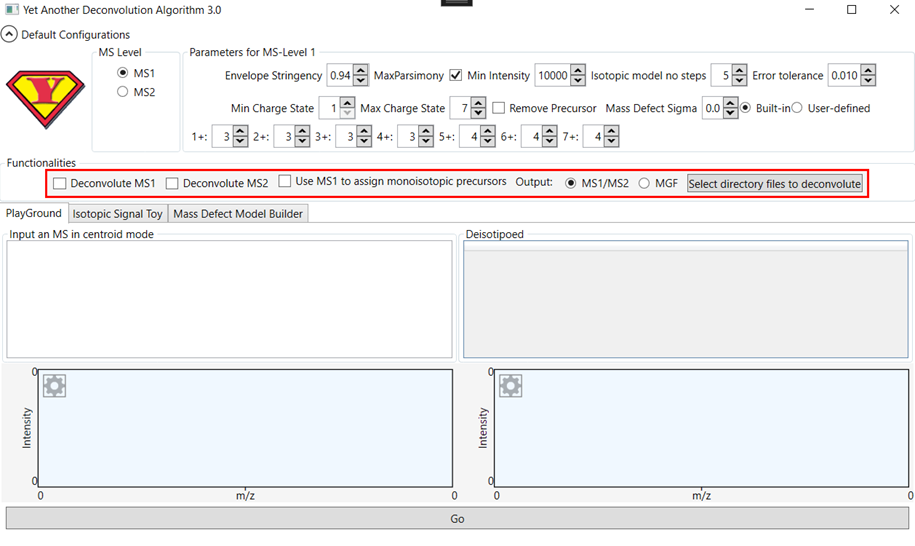
Figure 2 - Deconvoluting files in YADA 3.0
The next
step is actual deconvolution, which can be performed by clicking on the “Select
directory with .raw files to deconvolute” button. YADA 3.0 generates .ms1 and
.ms2 files containing the deconvoluted ions in the same directory.
If the “Use
MS1 to assign monoisotopic precursors” option is checked, YADA adds extra
Precursor ion information to the MS2 spectra, which is obtained from the
deconvolution results of the MS1 spectra. This strategy has led to improvements
in identification rates in PatternLab for
Proteomics.
4. Deconvolution Playground
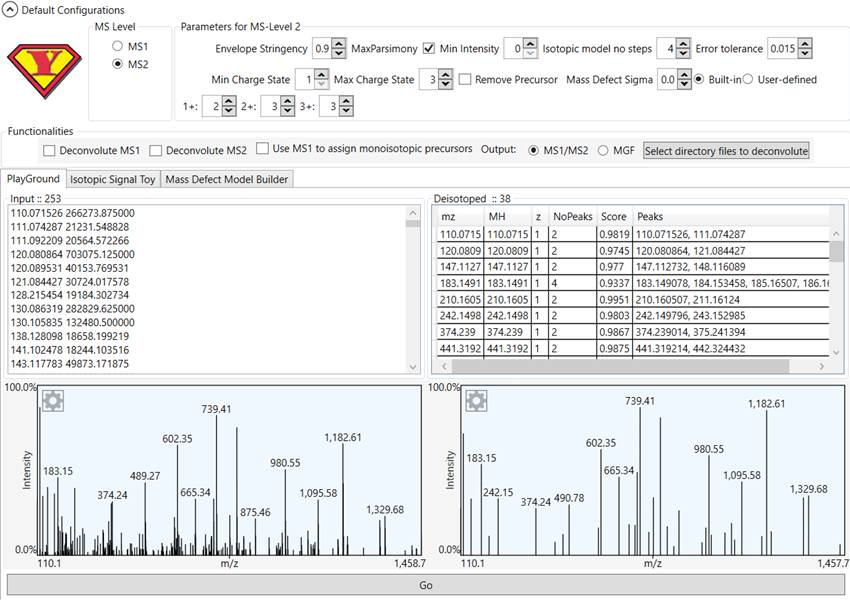
Figure 3 - YADA 3.0 Deconvolution Playground.
YADA 3.0
provides a tool for testing deconvolution parameters. For this, the user must
paste the spectrum ions in the .ms2 format (m/z + tab + intensity) and click
the “Go” button to apply deconvolution.
How to Use (Command Line Version)
1. Download
The command line
version of YADA 3.0 can be downloaded here. It
requires .NET 6.0 or higher to run, which can be downloaded here.
Sample
parameter .xml files can be downloaded here. These files contain the default
parameters for YADA 3.0 which should work well for most standard proteomic
samples.
An example
raw file can be found here.
2. Running
The
arguments for running the command line application are as follows:
YCore.exe ms_path
-m -ms1 ms1_xml_path -ms2 ms2_xml_path -i .raw -o .mgf
The
arguments are:
ms_path: path to either a file or a
directory containing ms files in the .raw, .ms1, .ms2,
or .mgf formats. If set as '\' will run on current
directory.
-ms1: Tag preceding the path for parameters xml for deconvolution
of MS1 spectra. If set to 'default', default parameters are used. If not set
deconvolution is not performed at MS1 level.
-ms2: Tag preceding the path for parameters xml for
deconvolution of MS2 spectra. If set to 'default', default parameters are used.
If not set deconvolution is not performed at MS2 level.
-i: Tag preceding the format for
input MS files. Supported formats are .raw, .ms1,
.ms2, and .mgf.
-o: Tag preceding format deconvoluted spectra are written to.
Supported formats are .ms1, .ms2, and .mgf.
-m: Add multiplex charges to outputs. Requires inputs to be in .raw format, or in .ms1 AND .ms2 formats.
Instructions
for running the command line version of YADA 3.0 can also be obtained in the
executable itself through the -help command:
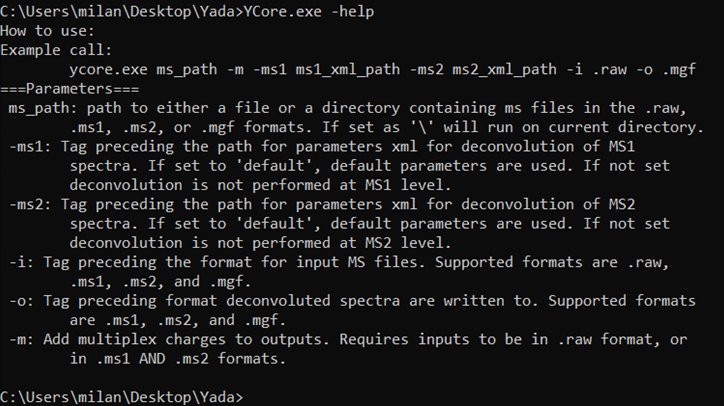
Figure 4 - Command line help command.
3. Parameters Description
For running
the command line application parameters may be provided as .xml files. An .xml
file must be provided for each MS level.
The .xml
file is as follows:
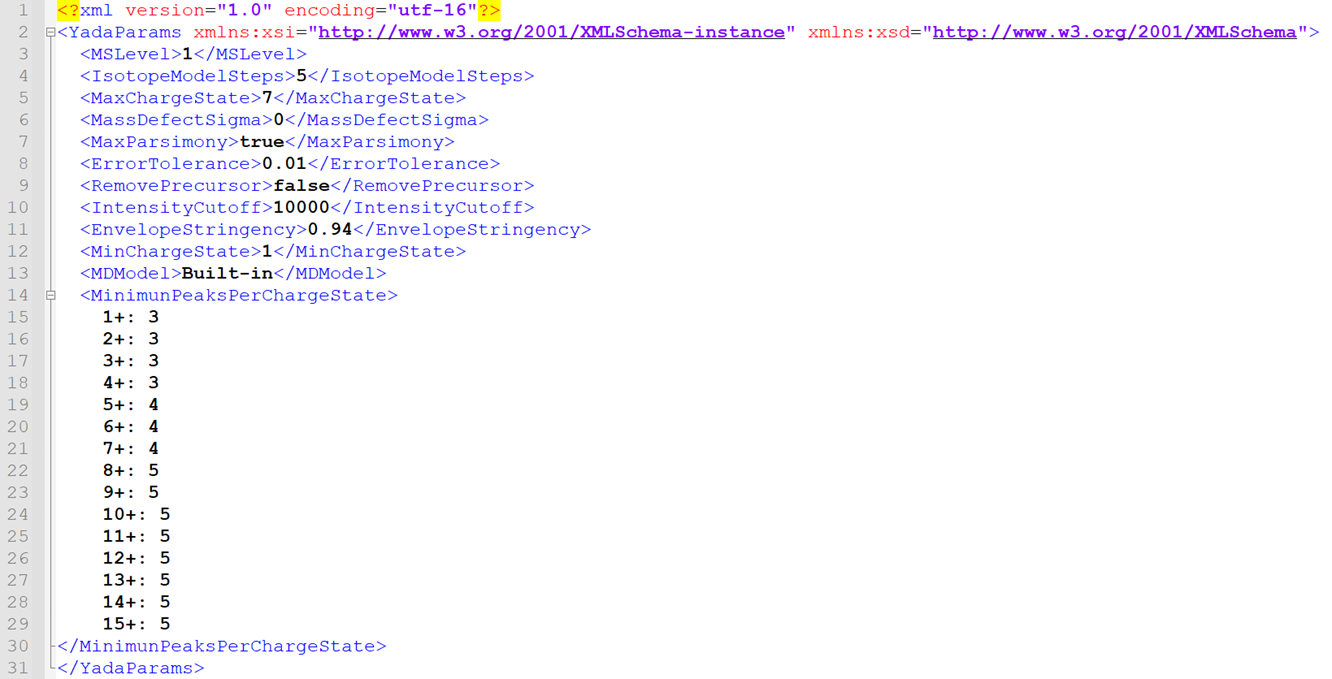
Figure 5 - Sample YADA 3.0 xml parameters file.
Where:
· MSLevel: Fragmentation level of spectrum.
For MS1 spectra it is 1, for MS2 spectra it is 2, and so on.
· Isotopic Model Steps: Number of
isotopes to be modelled for each ion.
· MassDefectSigma: Mass error allowed in mass defect
calculations. Set to zero to not consider mass defect.
· MaxParsimony: Whether to allow an ion to belong
to two different isotopic envelope. If checked, the
lower scoring envelope for each ion is ignored.
· Error Tolerance: Mass error allowed.
Measured in Daltons.
· RemovePrecursor: Whether to remove ions matching the
precursor from MS2 spectra.
· IntensityCutoff: Minimum intensity required for ions
to be considered for isotopic envelopes.
· Envelope Stringency: Minimum isotopic
envelope similarity score required to accept an envelope.
· Min Charge State: Minimum charge
state to consider in the spectrum.
· MDModel: Whether to use the Built-In mass
defect error model, or provide a new one. Keep as
‘Built-in’.
· Remove Precursor: If selected,
envelopes matching the precursor mass will be ignored.
· MinimunPeaksPerChargeState: Minimum number of ions required to
accept an isotopic envelope for different charge states..
Optimal Parameters
Optimal
parameters for running YADA 3.0 may change depending on the target dataset.
For
bottom-up and middle-down proteomics we recommend:
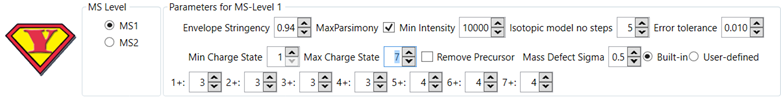
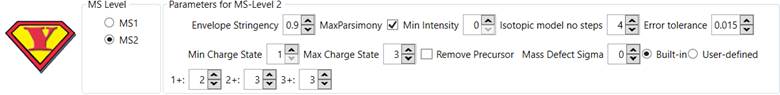
Figure 6 - Recommended YADA 3.0 parameters for bottom-up and
middle-down proteomics.
For top-down
proteomics we recommend:

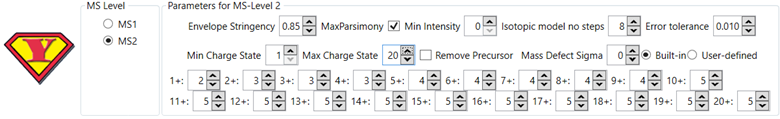
Figure 7 - Recommended YADA 3.0 parameters for top-down
proteomics.
These parameters are also supplied as .xml files here.