
RawVegetable’s Tutorial
PROTOCOL
RawVegetable requires Thermo mass spectrometer RAW
files, *.mzML or Agilent
files for the main feature to function. The chromatography reproducibility
feature requires PatternLab for proteomics’ *.xic or *.plp files or a SIM-XL
output file. In order to view identifications along the chromatogram, a SIM-XL
file is required.
Example dataset can be downloaded here.
Extended information on how the modules
work can be found here.
HOW TO USE
1.
Installing RawVegetable
The latest version of the RawVegetable software is available here. The
software requires version 4.7.2 or higher of the .NET framework, which can be
downloaded here, if
it is not already installed.
For reading Thermo RAW files,
the MSFileReader must be installed. Download it by
creating an account at Thermo®, then logging and choosing Utility
Software.
2.
Charge State Chromatogram
2.1. Select
the chosen RAW or mzML files by clicking on the “Load Data” button
(which can be found in the “Files” menu).
To open an Agilent file, click on “Load Agilent File” (Figure 1). You
can also simply drop the files you wish to analyze.
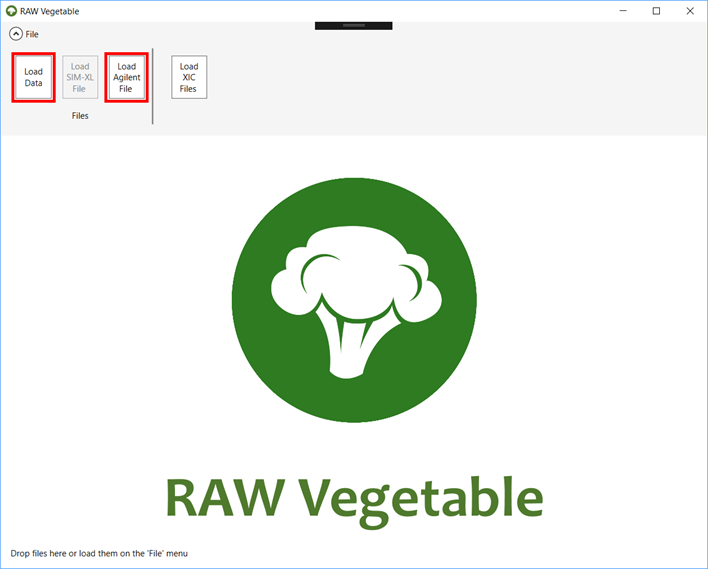
Figure
1 - Starting RawVegetable
Screen.
2.2. First
you will have to choose between extracting the chromatogram as Total Ion
Current (TIC) or Base Peak Only, as shown in Figure 2.
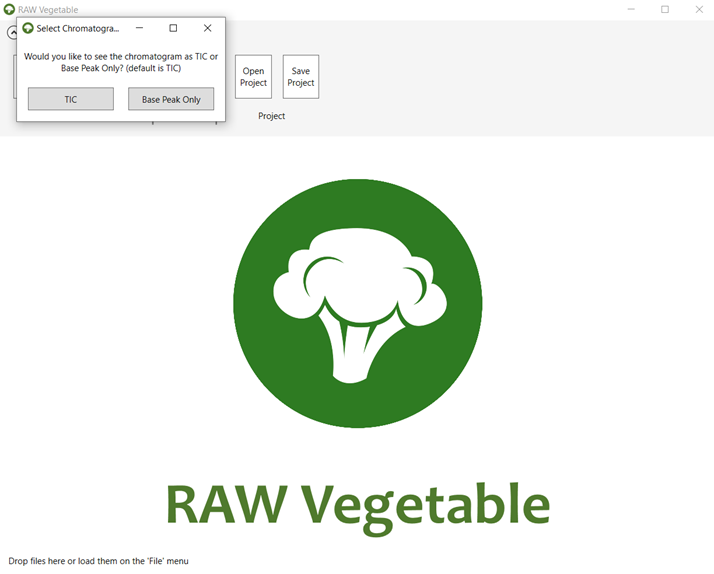
Figure 2.
2.3. After
the file loads, the screen will be as shown in Figure 3.
Information such as the number of MS1 and MS2 scans can be seen on the “General Information” tab.
2.4. To see
all the files loaded and choose which ones to show on the viewer, click on the “Open Files” menu (Figures 3 and 4).
Simply click on the checkbox to view/hide a chromatogram.
2.5. In
order to see the charge-specific chromatograms, click on the “Deconvolute” button (Figure 3). This
process will deconvolute all the files that are checked in the “Open Files” menu (Figure 4) and
can take minutes to conclude according to the size of the files.
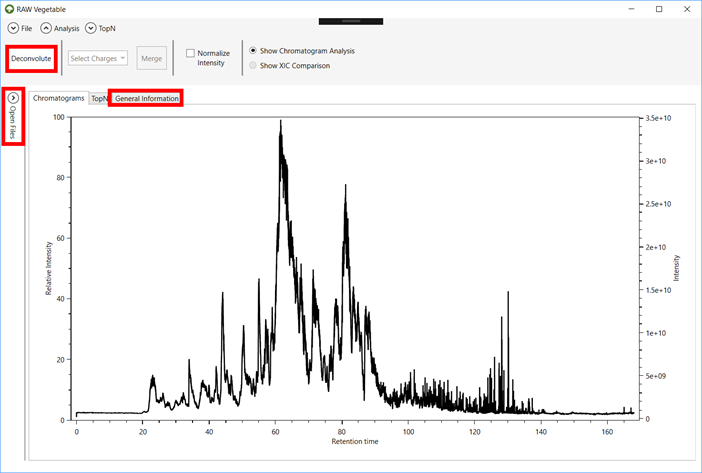
Figure 3.
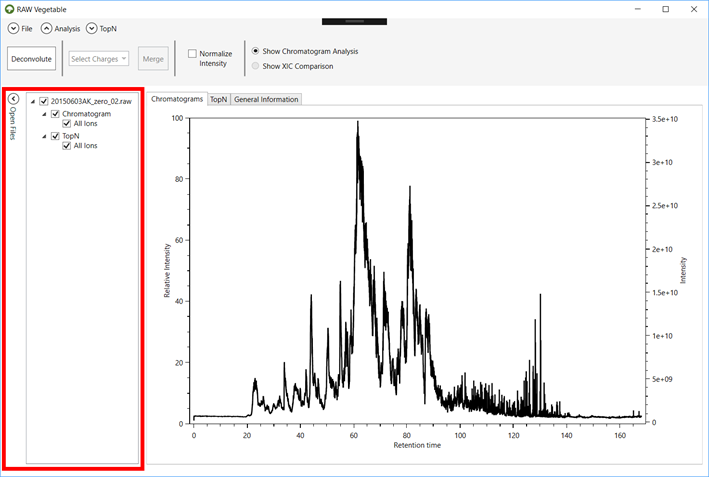
Figure 4.
2.6. Once
the deconvolution is complete, select the charge you want to view on the “Open Files” menu (Figure 5). You
can identify the chromatograms by hovering the mouse on top of it.
2.7. To see
all chromatograms as relative intensities (normalized values), click on the “Normalize Intensity” checkbox (Figure 5).
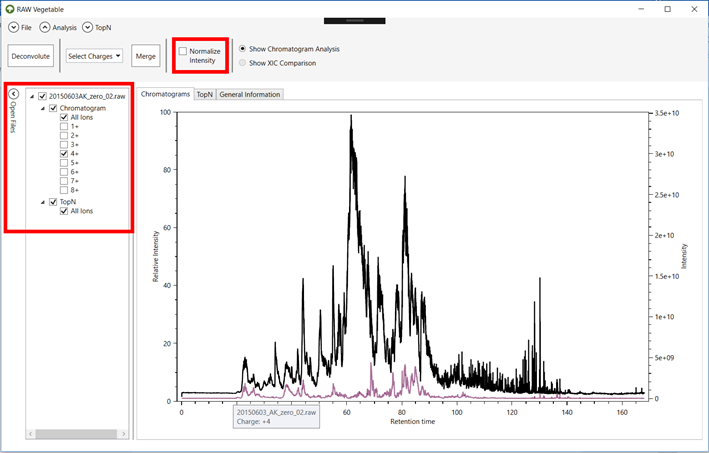
Figure 5.
2.8. If you
want to see more than one charge as a single chromatogram, select the charges
on the combo box shown in Figure 6 and
click the “Merge” Button. The merged chromatogram will now be
shown on the “Open Files” menu.
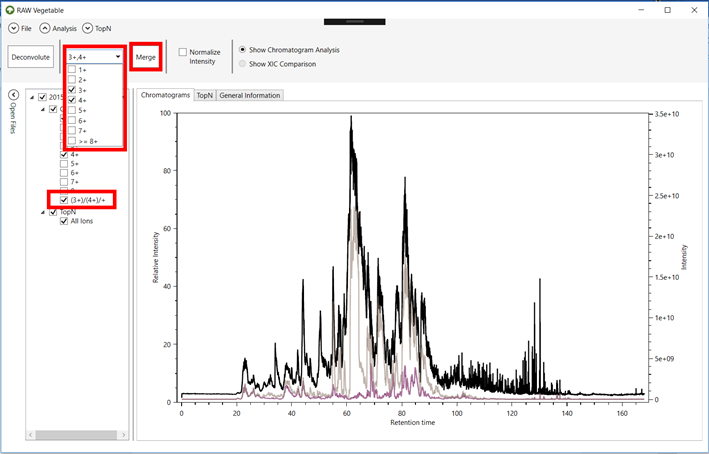
Figure 6.
2.9. If you
have a SIM-XL output file and wish to see where the cross-links were
identified, click on the “File” menu
and on the “Load SIM-XL File” button (Figure 7).
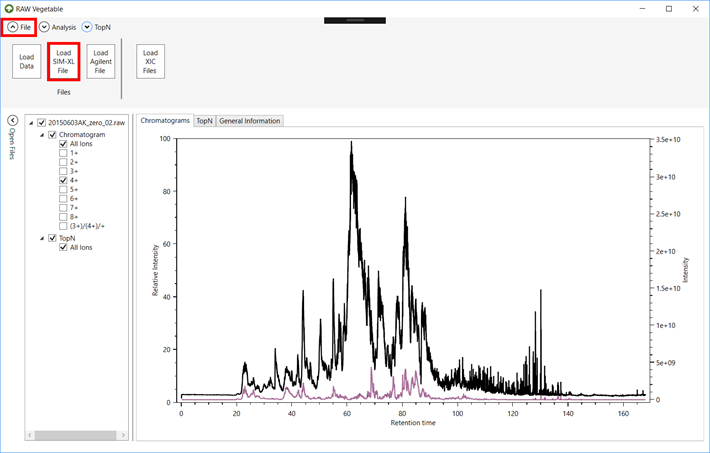
Figure 7.
2.10. After
the file loads, the “SIM-XL” menu
will appear on top, where you can filter the identifications by score and link
type. To see the cross-links, click on the “Show Identifications” button
(Figure 8).
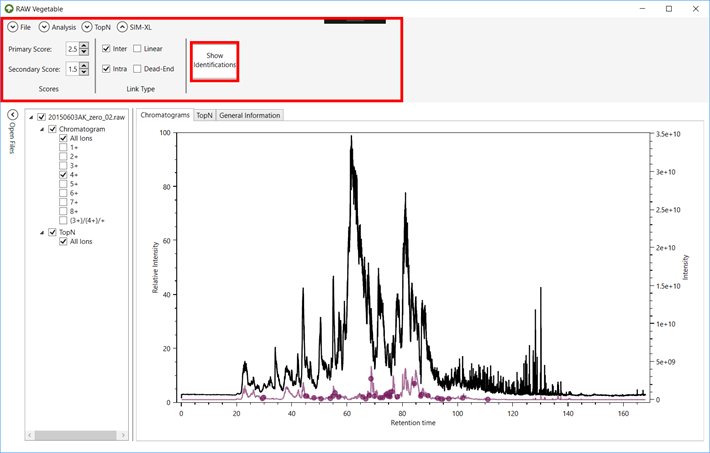
Figure 8.
3.
TopN Density Estimation
3.1. To see
the TopN density estimation, click on the “TopN” tab after loading the files (Figure 9).
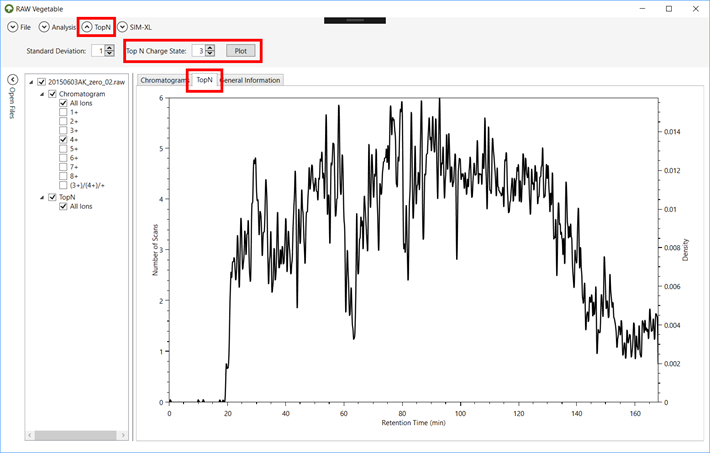
Figure 9.
3.2. To see
the density estimation only of precursors of a specific charge, go to the “TopN” menu, select the charge and
click on “Plot” (Figure 9). This
might take a while according to the number of MS2 scans acquired.
3.3. Once
the density estimation has been calculated, the curve will appear on the TopN viewer and you can view/hide it by clicking on the
checkbox under the “TopN” item
for each file on the “Open Files” menu (Figure 10).
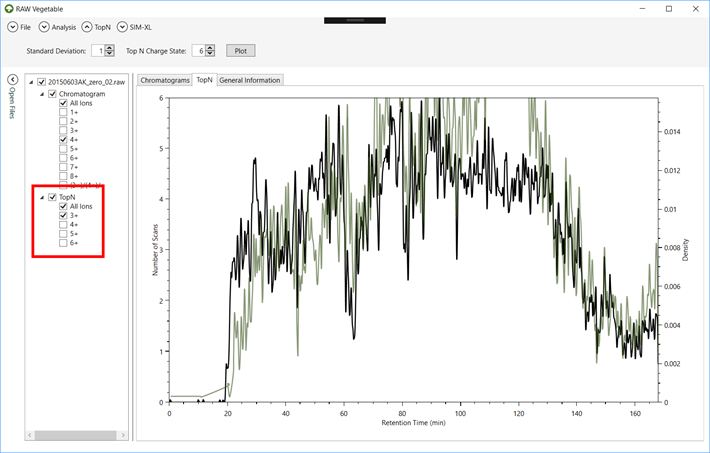
Figure 10.
4.
Chromatography
reproducibility
4.1. Load
either PatternLab for proteomics’ *.xic or *.plp files, or SIM-XL
output files (*.simxlr) by going to the “File” menu
and clicking on “Load XIC Files” (Figure 11).
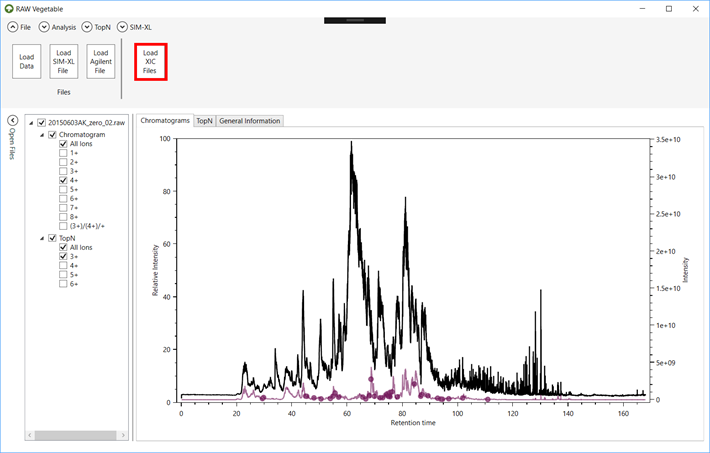
Figure 11.
4.2. You can
see the heatmap generated by the comparison of all files on the right, or you
can choose two files to compare on the left side (Figure 12). It
is also possible to choose to look for all peptides instead of unique or common
ones, by clicking on the checkboxes on the left.
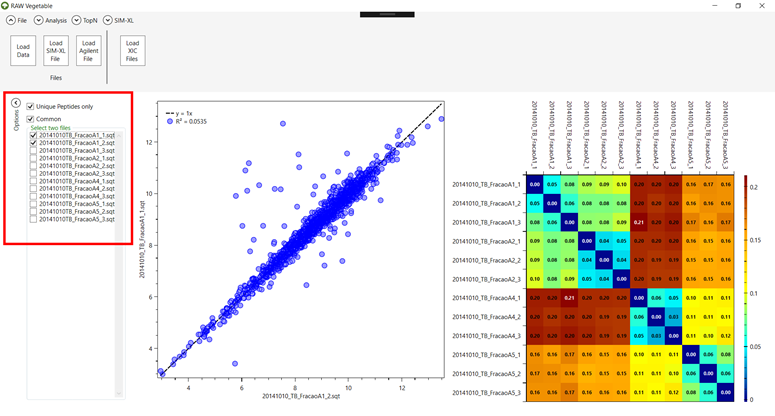
Figure 12.
4.3. To go
back to seeing the chromatography analysis, go to the “Analysis” menu and click on the “Show Chromatogram Analysis” checkbox (Figure 13).
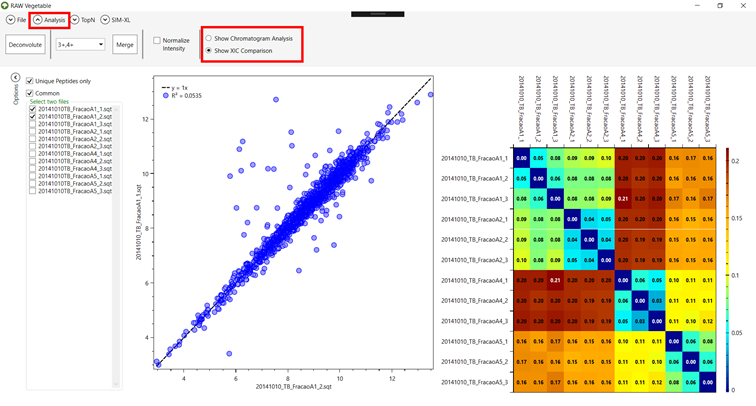
Figure 13.