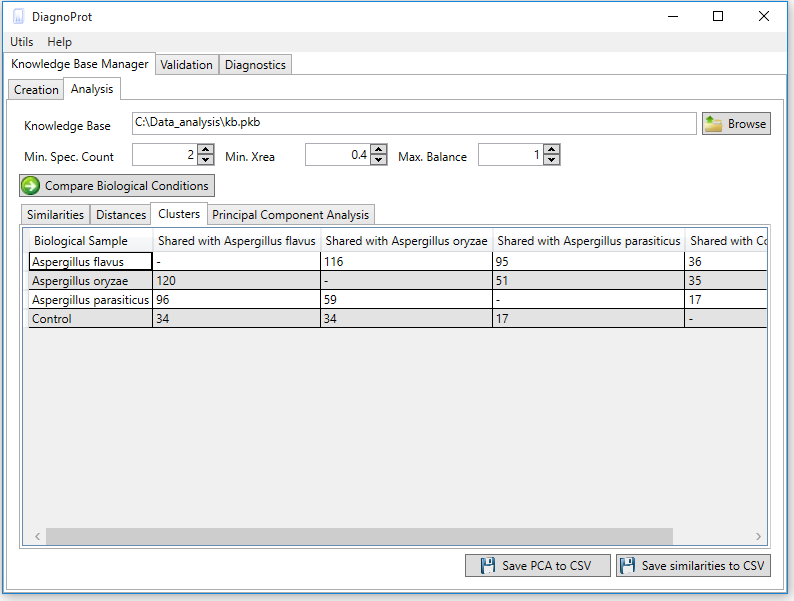
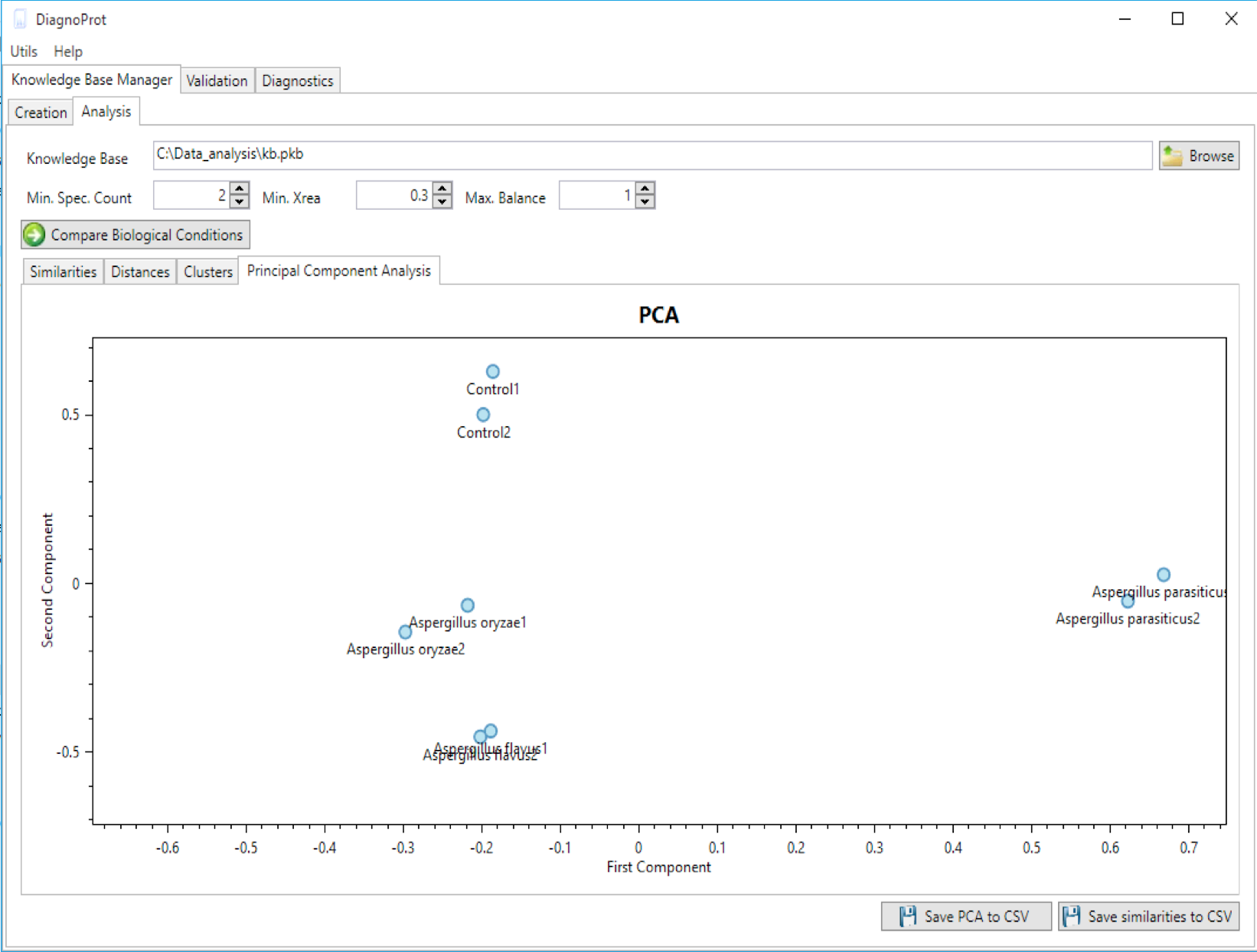
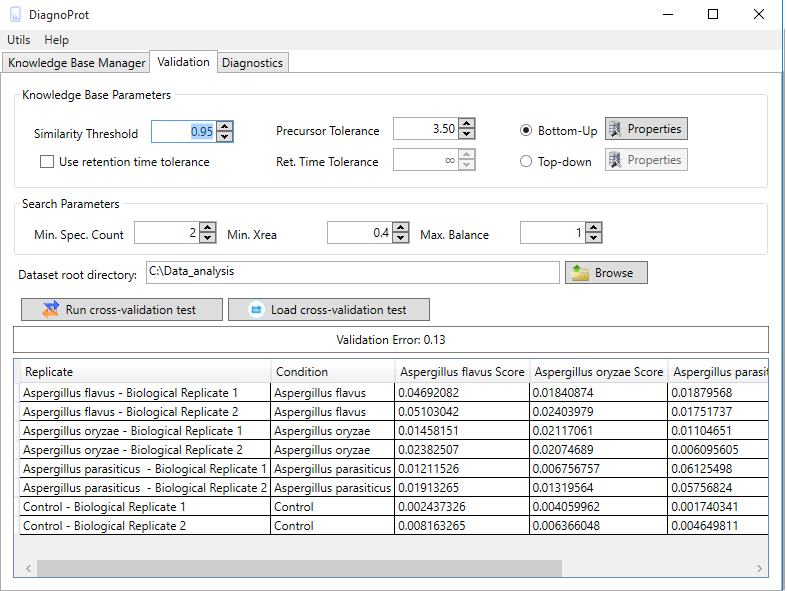
Welcome to DiagnoTop
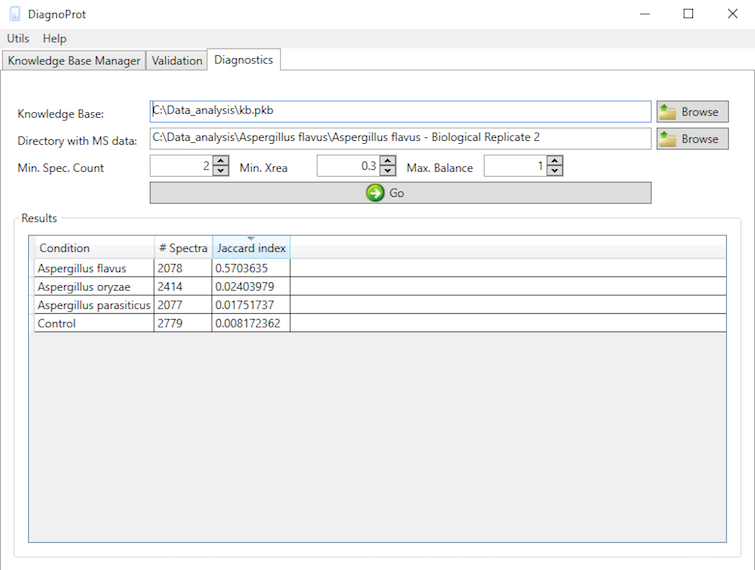
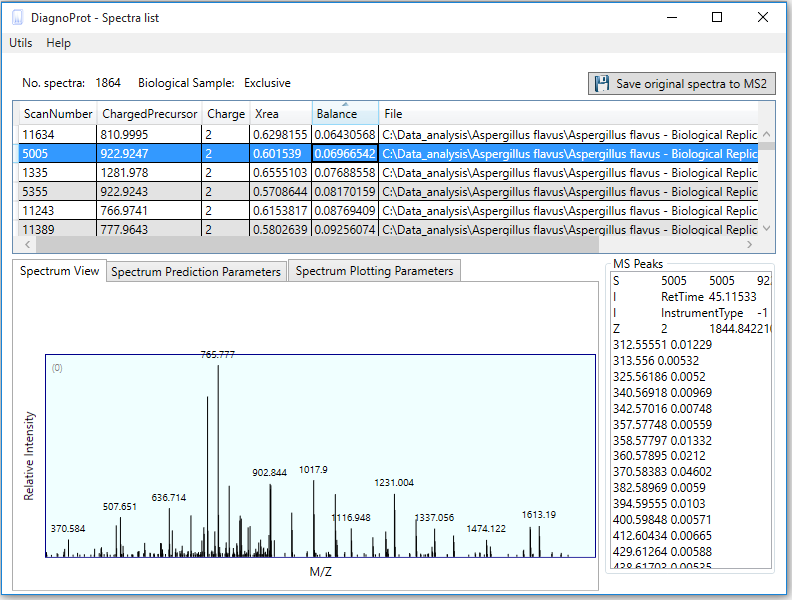
Pathogen identification is crucial to confirm bacterial infections and guide antimicrobial therapy. Although MALDI-TOF mass spectrometry (MS) is a powerful tool for rapid microbial identification, some bacteria remain challenging to identify. We recently showed that top-down proteomics (TDP) could be used to discriminate closely related enterobacterial pathogens (E. coli, Shigella, and Salmonella) that are indistinguishable with MALDI-TOF MS. This diagnostic relies on the identification of specific proteoforms for each species through a database search. However, microbial proteomes are often poorly annotated, which complicates the large-scale identification of proteoforms and leads to many unidentified high-quality mass spectra. Here, we describe a computational approach., called DiagnoTop, where we use DiagnoProt together with Top-Down Garbage Collector, to shortlist discriminative spectral clusters found in TDP datasets that can be used for microbial diagnostics without database search. This pipeline opens new perspectives in clinical microbiology and biomarker discovery using TDP.
 André R F Silva, Diogo B Lima, Alejandro Leyva,
Rosario Duran, Carlos Batthyany, Priscila F Aquino, Juliana C Leal,
Jimmy E Rodriguez, Gilberto B Domont, Marlon D M Santos, Julia Chamot-Rooke,
Valmir C Barbosa, and Paulo C Carvalho,
“DiagnoProt: a tool for discovery of new molecules by mass spectrometry”,
Bioinformatics (2017) 33 (12): 1883-1885.
André R F Silva, Diogo B Lima, Alejandro Leyva,
Rosario Duran, Carlos Batthyany, Priscila F Aquino, Juliana C Leal,
Jimmy E Rodriguez, Gilberto B Domont, Marlon D M Santos, Julia Chamot-Rooke,
Valmir C Barbosa, and Paulo C Carvalho,
“DiagnoProt: a tool for discovery of new molecules by mass spectrometry”,
Bioinformatics (2017) 33 (12): 1883-1885.