What is PatternLab?
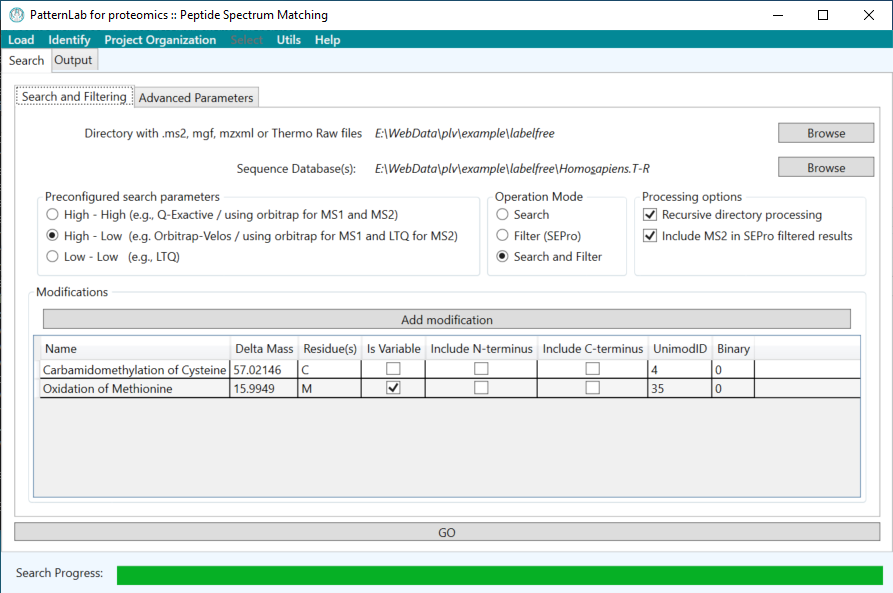
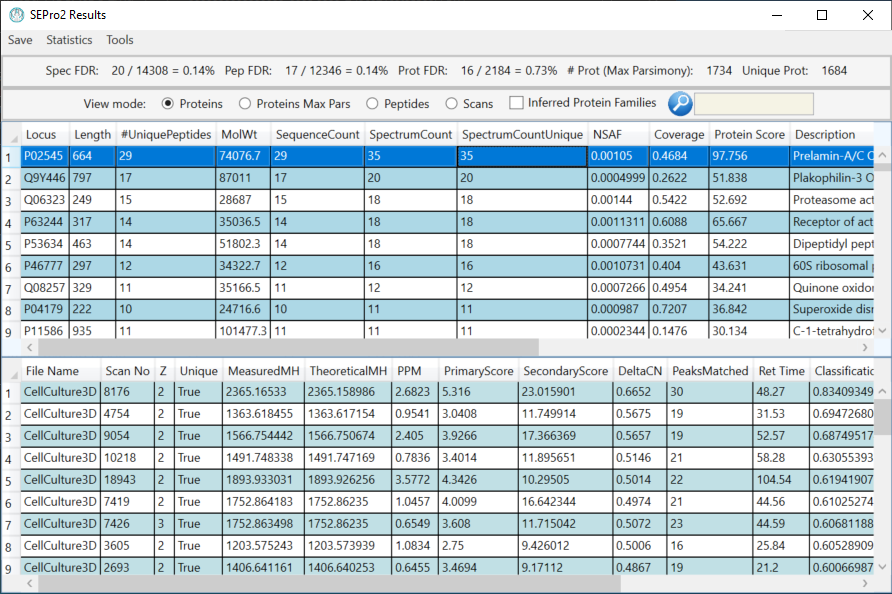
PatternLab for Proteomics is an integrated computational environment for analyzing shotgun proteomic data. PatternLab contains modules for formatting sequence databases, performing peptide spectrum matching, statistically filtering and organizing shotgun proteomic data, extracting quantitative information from label-free and chemically labeled data, performing statistics for differential proteomics, displaying results in a variety of graphical formats, performing similarity-driven studies with de novo sequencing data, and analyzing time-course experiments. Here, we present PatternLab for Proteomics V, which closely knits together all these modules in a self-contained environment, covering the principal aspects of proteomic data analysis as freely available and easily installable software.PatternLab is user-friendly, simple and provides a graphical user interface.
Please cite:
doi> “Integrated analysis of shotgun proteomic data with PatternLab for proteomics 4.0”, Nature Protocols (11), 102-117, 2016.